Conversation
There was a problem hiding this comment.
@guiwitz this is BEAUTIFUL. Gorgeous, fantastic, amazing, well written, and inspiring. You clearly spent a ton of time on this and I'm absolutely LOVING this contribution. I don't have too many comments because I really like the organization and flow.
Also... Holy smokes @guiwitz I actually had a panic attack. 😱 Have you actually tried ndevio? It's certainly mature enough that I share it with folks, but to put it in here like this is both an honor and so intimidating. I am a big believer in it, so getting eyes on it would be great lol. I guess the main flaw with it is that it's pypi only at the moment. It looks like I'd be able to add it to conda-forge with bioio-ome-tiff and bioio-ome-zarr (and the option for bioio-lif) which would at least be a start! I will work on it this week (regardless!). Unfortunately, I don't know of another "catch-all" reader that works with modern napari, so that might be the one to go with.
Would be good to get feedback from others.
Also, this might be a good file to mention https://github.com/napari/napari-tiff . I havne't worked on this much, so hopefully others can describe its value better than I can.
I added dependencies to
pixi.toml, but being a complete beginner with it, someone should check if that's correct (especially given the multiple sections with dependencies). Also those dependencies might be missing when not using pixi?
For this, we have the unfortunate problem that we would need to add the dependencies to napari's pyproject -- the full build workflow on CI will fail right now (if we trigger it with the bot). https://github.com/napari/napari/blob/326d0144b22b7808cd16dfeb49c48590d94869d9/pyproject.toml#L276-L304
It's one of those complicated issues with how we have things organized, sadly.
But we can test with pixi though locally! i.e. pixi run docs-live. For some reason it seems like the code blocks aren't showing outputs, despite seeing C:\Users\timmo\Documents\GitHub\napari-docs\docs\getting_started/open_images.md: Executing notebook using local CWD [mystnb]... so I'll try to look into it
Good news too, the additional dependencies don't add too much, which is good and should be easily maintainable:
+ P aicspylibczi 3.3.1
+ P aiobotocore 3.1.0
+ P aioitertools 0.13.0
+ P bioio 3.2.0
+ P bioio-base 3.2.0
+ P bioio-czi 2.4.2
+ P bioio-imageio 1.3.0
+ P bioio-ome-tiff 1.4.0
+ P bioio-ome-zarr 3.2.1
+ P bioio-tifffile 1.3.0
+ P botocore 1.42.19
+ P cmake 4.2.1
+ P elementpath 5.1.0
+ P imagecodecs 2026.1.1
+ P jmespath 1.0.1
+ P kerchunk 0.2.9
+ P macro-kit 0.4.10
+ P magic-class 0.7.20
+ P napari-meshio 0.0.2
+ P napari-skimage 0.6.0
+ P nbatch 0.0.4
+ P ndev-settings 0.4.0
+ P ndevio 0.7.0
+ P ome-types 0.6.3
+ P pydantic-extra-types 2.11.0
+ P pylibczirw 5.1.1
~ P pyqt5-sip 12.17.2 -> 12.18.0
+ P s3fs 2026.1.0
+ P semver 3.0.4
+ P ujson 5.11.0
+ P validators 0.35.0
+ P xmlschema 4.3.0
+ P xmltodict 1.0.2
+ P xsdata 25.7
docs/getting_started/open_images.md
Outdated
| nbscreenshot(viewer, alt_text="napari viewer showing a multi-channel CZI image opened via ndevio of Ostrinia nubilalis cells.") | ||
| ``` | ||
|
|
||
| On top of just allowing to open file formats, some plugins also provide additional tools, for example to explore metadata, offer import options, etc. The ndevio plugin for example provides widgets to export data and explore basic metadata via the `Plugins -> ndevio -> I/O utilities` menu. Here we see that valuable information about pixel size was recovered from the czi file metadata: |
There was a problem hiding this comment.
Once we release napari-metadata, I think this would be a good spot to also expose metadata (and would be my preference). At that point, I will likely reduce my I/O Utilities widget to be much slimmer and prioritize napari-metadata
There was a problem hiding this comment.
I can also remove that section. But I find it useful to show that some readers offer more than just opening images. It's the case also e.g. for the napari-czitools (but I didn't want to have that plugin as a dependency).
| nbscreenshot(viewer, alt_text="napari viewer showing a multi-channel TIFF image of E. coli cells opened via ndevio.") | ||
| ``` | ||
|
|
||
| In the case of badly interpreted dimensions, one can also reshape the data using certain plugins. For example the [napari-skimage](https://napari-hub.org/plugins/napari-skimage.html) plugin provides a tool to reshape layers via the `Plugins -> napari skimage -> Axis operations` menu. Here we use the `swap axes` function to exchange the X and Y axes: |
There was a problem hiding this comment.
This will be another spot we can refer to napari-metadata
Co-authored-by: Tim Monko <timmonko@gmail.com>
Co-authored-by: Tim Monko <timmonko@gmail.com>
Co-authored-by: Tim Monko <timmonko@gmail.com>
|
Regarding dependencies: is there an explanation anywhere on the "machinery" used to build docs? It's not clear to me why the dependencies have to be added to napari. |
|
I don't think there's anything beyond https://napari.org/stable/tutorials/fundamentals/installation.html#using-constraints-files . Unfortunately since we're using the two repos it's a bit convoluted, but this allows us to do
and have the docs dependencies follow the general constraints required for reliably building napari itself. |
 DragaDoncila
left a comment
DragaDoncila
left a comment
There was a problem hiding this comment.
This is awesome @guiwitz 🤩 ! I left a couple of comments, but I think once we figure out the CI issues this is ready to go in. I mentioned to Tim earlier today that it would be great to expand a bit on the different File -> Open options (File, File(s), File(s) as stack, Folder...), but that should come in a separate PR imo.
Co-authored-by: Draga Doncila Pop <17995243+DragaDoncila@users.noreply.github.com>
Co-authored-by: Draga Doncila Pop <17995243+DragaDoncila@users.noreply.github.com>
Co-authored-by: Draga Doncila Pop <17995243+DragaDoncila@users.noreply.github.com>
Co-authored-by: Draga Doncila Pop <17995243+DragaDoncila@users.noreply.github.com>
|
Woohoo! Once we have the dependencies in we can start building this and looking at it. We discussed this at last weeks docs meeting that we want to reduce the size of the pooch retrieved images because 1) it will slow down CI and 2) we are then pinging someone's servers constantly. It's really nice that you were able to get these images from a place that isn't Zenodo, because we've had difficulty from them recently. Do you have any smaller data that we could use (ideally <5MB or so)? We are also happy to help locate data for this because we know how challenging it can be to locate stuff. Also, napari-metadata is released, so we can rethink how to add that here. But I don't want to slow down this PR any more than we have, so we should perhaps save it for a follow-up :) (unless you want to work on it here) |
# References and relevant issues The [docs PR](napari/docs#907) about opening images requires three plugin dependencies for the document to be generated. # Description This PR adds three docs dependencies to fix the issue above. --------- Co-authored-by: Melissa Weber Mendonça <melissawm@gmail.com> Co-authored-by: Draga Doncila <ddon0001@student.monash.edu> Co-authored-by: Tim Monko <timmonko@gmail.com>
Co-authored-by: Tim Monko <timmonko@gmail.com>
|
Grrr, our constraints process failed again: I tried to look into supressing that warning, but it wasn't really working.
So I think for now it's best to just set upper pin on sphinx-external-toc. |
|
@napari-bot make html |
|
OK, here is the pinning PR: napari/napari#8591 |
This information comes from napari/napari/pyproject.toml docs dependency-group
…into open-images
|
Here is a quick summary of the why of my changes:
Again thank you so so so so so much for this work @guiwitz, it might look like I did a lot, but I mostly just built on your great PR and moved things around 🥳 🍻 |
|
@napari-bot make html |
|
@napari-bot make html |
|
@TimMonko looks like there's a legitimate docs build issue here. I've had a read through the content and I think it's good otherwise! |
|
Changes look great @TimMonko, especially the new napari-metadata section! I had a look at the issue, which I only understood after creating a fresh local environment: we demo how to use |
# References and relevant issues napari/docs#907 and #50 # Description This adds a test zarr that gets seen as a directory in Windows, and sets the manifest to accept directories. @guiwitz was unable to get zarrs working with ndevio. I realized that one reason is that sometimes .zarr gets interpreted locally as a file, and sometimes as a directory. Mistakenly, I only ever tested .zarr that were interpreted as a file, so my napari.yaml manifest ended up not accepting file directories.
 jni
left a comment
jni
left a comment
There was a problem hiding this comment.
Thanks @DragaDoncila for reminding me to look at this PR. I love all the autogeneration going on here! Some comments:
- I think images (e.g. screenshots) in raw.github should be in
_static - I think data that is in raw.github should be moved to Cloudflare. Once we agree on a strategy I can move them.
- Other files would also be good to have under our control, e.g. someone's psu.edu page, so maybe we can say what the source is but store a copy in Cloudflare and build from that. Thoughts? (Bioimage Archive maybe we can just leave, that seems less temporary.)
- I kinda disagree with @TimMonko that the new dependencies are "small", and his list makes me kinda nervous, but I think it would be hairy to reduce them, so let's go ahead if we can get CI working.
- let's set the download directory at the top of the file (and make it if necessary), defaulting to, for example,
~/Desktop/napari-docs-build, not~. Files in~are not visible on most OSs so the building the docs locally would be adding cruft to people's computer that they would have a hard time seeing.
|
|
||
| With the release of napari 0.7.0, [napari-metadata](https://napari-hub.org/plugins/napari-meta) is included with the bundle, conda-forge and with `pip install "napari[optional]"` (or `napari[all]`) installations. This plugin provides a widget to view and edit the metadata of layers. This can be useful to check if the metadata was correctly read, and to edit it if necessary. | ||
|
|
||
| 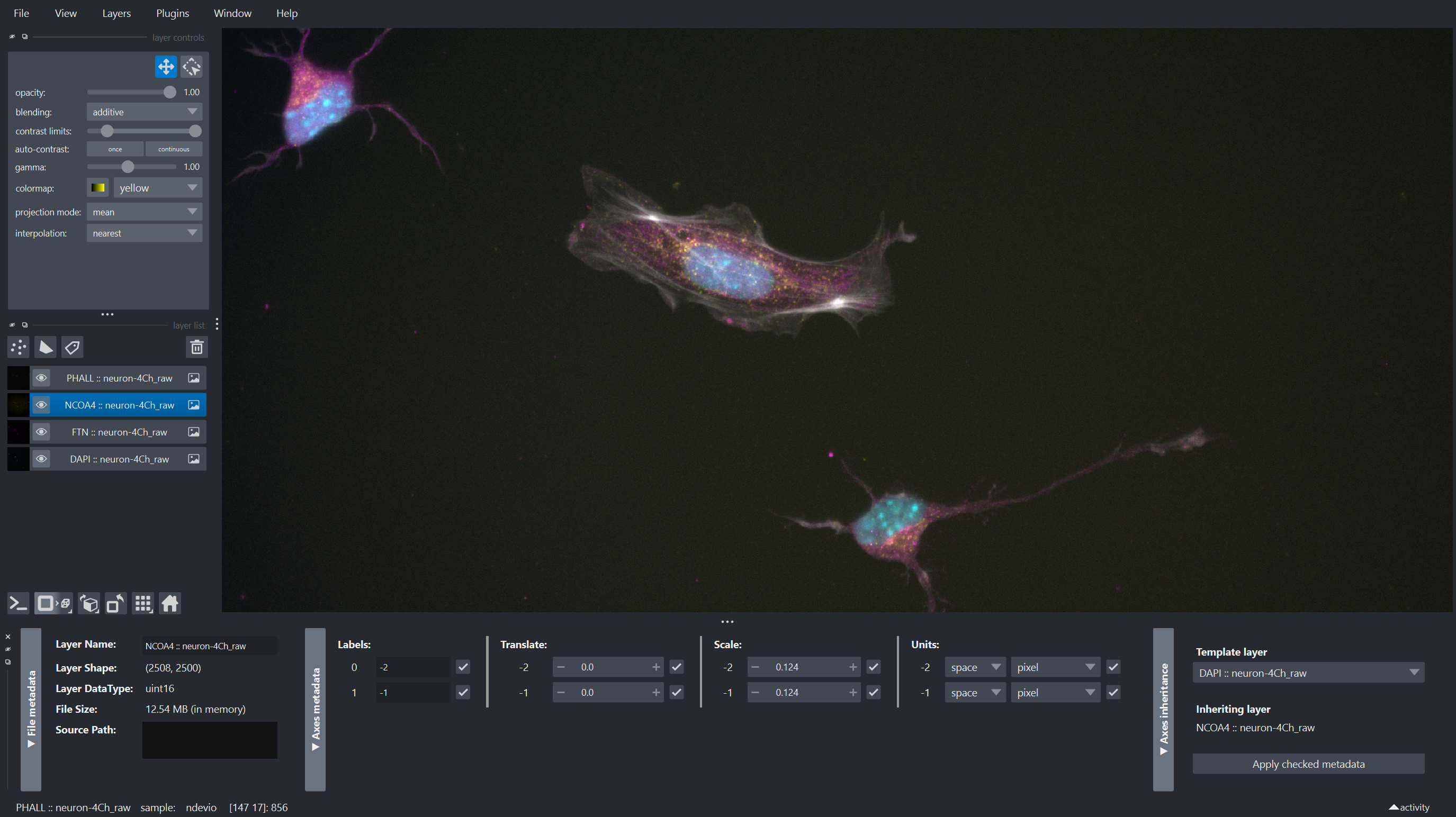 |
There was a problem hiding this comment.
This should be added to _static. We decided a while back not to rely on raw.githubusercontent links in the docs, as they could one day be throttled.
There was a problem hiding this comment.
This is from the napari-metadata plugin not the docs. Should we duplicate that file?
There was a problem hiding this comment.
I now modified the download path to ~/Desktop/napari-docs-build
|
I now removed the dependency to the CZI reader and I'm opening a v5 ome.zarr file which ndevio supports by default. This is possible thanks to a recent fix to ndevio by @TimMonko, so it will only work build correctly once Tim makes a new release of ndevio. The idea is just to show something else than a tiff file without depending on another install, but if someone has a better idea, let me know. I changed to |
|
One last question to the docs people. I see references to both the plugin section from the docs and the napari-plugin-manager docs (which I didn't know existed). Is that ok, should we only point to the manger? Are all the plugin manager docs going to move to that new location? |

References and relevant issues
Improving the section about images was discussed at the napari hackaton and in a more general PR about docs reorganisation.
Description
The PR expands and modernizes the previous version. The main improvement is a more detailed section about using reader plugins to import formats not natively supported by napari. In more details, the section now:
All the examples use downloadable data (from openmicroscopy and the BioImage Archive) and are generated, not screenshots. As discussed in #working-group-documentation > Capturing images, this implies that plugins are required to build the documentation. I added dependencies to
pixi.toml, but being a complete beginner with it, someone should check if that's correct (especially given the multiple sections with dependencies). Also those dependencies might be missing when not using pixi?Depends on napari/napari#8581